SMRT® Cell: Consumable substrates comprising arrays of zero-mode waveguide nanostructures.
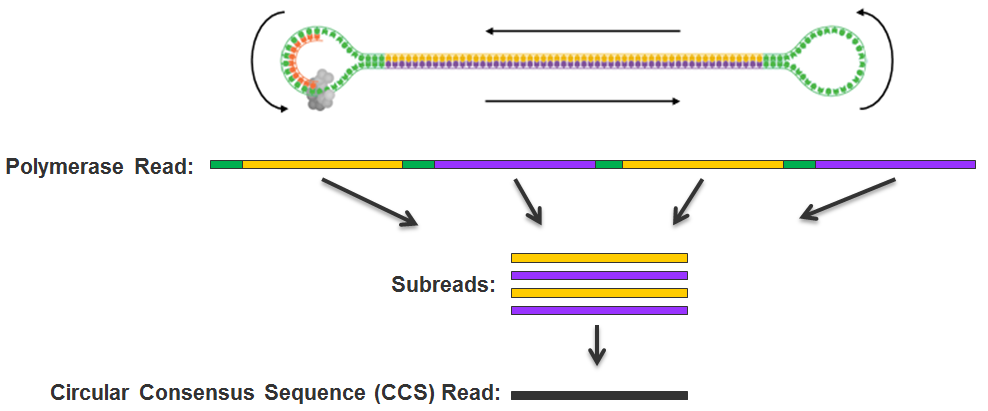
Adapters: Exogenous nucleic acids that are ligated to a nucleic acid molecule to be sequenced. For example, SMRTbell™ adapters are hairpin loops that are ligated to both ends of the double stranded DNA insert to produce a SMRTbell™ sequencing template. When adapter sequences are removed from a CCS read, the read is split into multiple subreads.
The wells and SMRT Cells to include in the sequencing run.
The collection and analysis protocols to use for the selected wells and cells.

circular consensus (CCS) read: The consensus sequence determined using subreads taken from a single ZMW. This is not aligned against a reference sequence. In contrast to Reads of Insert, CCS reads require at least two full-pass subreads from the insert.
read of insert: Represents the highest quality single sequence for an insert, regardless of the number of passes. For example, if your template received one-and-a-half subreads, that information will be combined into a Read of Insert. CCS is an example of a special case where at least two full subreads are collected for an insert. Reads of Insert give the most accurate estimate of the length of the insert sequence loaded onto a SMRT® Cell. For long templates, Reads of Insert may be the same as Polymerase Reads.
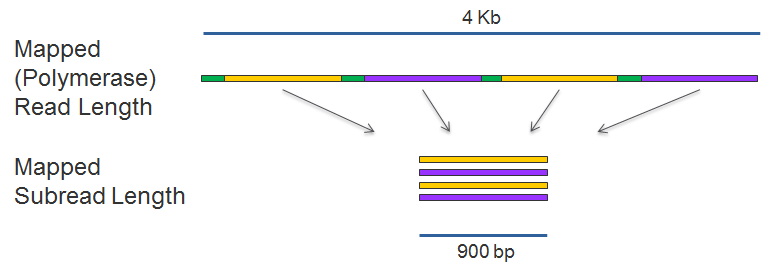
mapped polymerase read length: The total number of bases along a read from the first adapter or aligned subread to the last adapter or aligned subread. Approximates the sequence produced by a polymerase in a ZMW.
mapped subread length: The length of the subread alignment to a target reference sequence. This does not include the adapter sequence.
polymerase read length: The total number of bases produced from a ZMW after trimming. This may include the adapter sequence.
raw read trimming: Extraction of high quality regions from an unfiltered read. Trimming of an unfiltered read produces a polymerase read.
Read Quality ≥ .75 (as of SMRT Analysis v2.1)
Read Length ≥ 50 bases
circular consensus accuracy: Accuracy based on multiple sequencing passes around a single circular template molecule.
consensus accuracy: Accuracy based on aligning multiple sequencing reads or subreads together, optionally with a reference sequence.